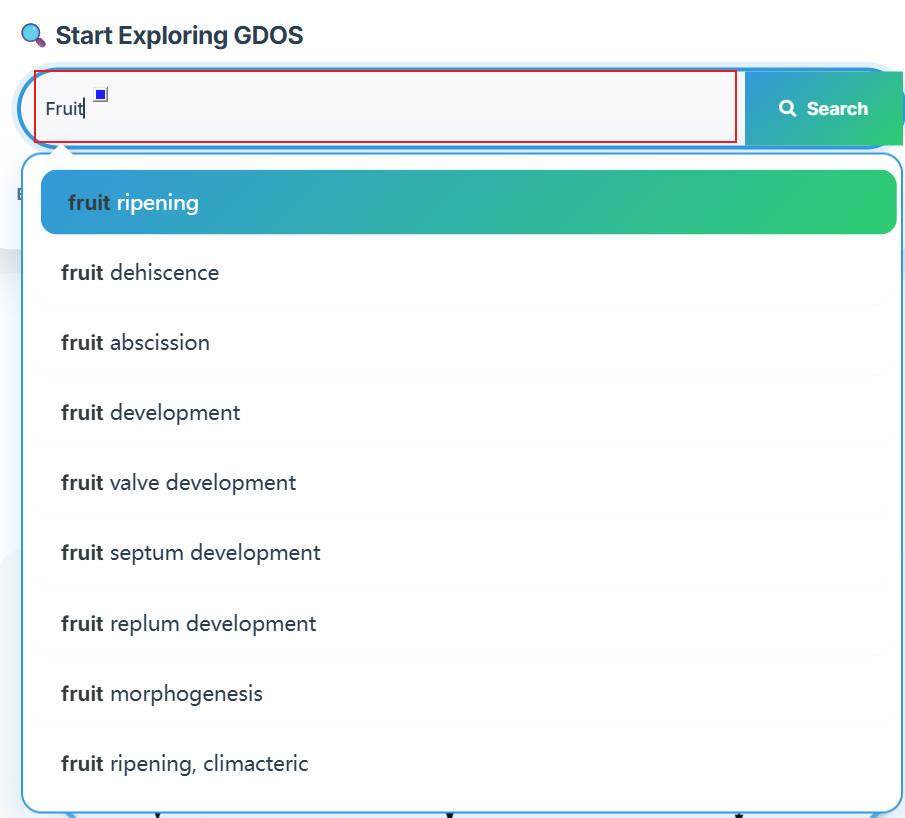
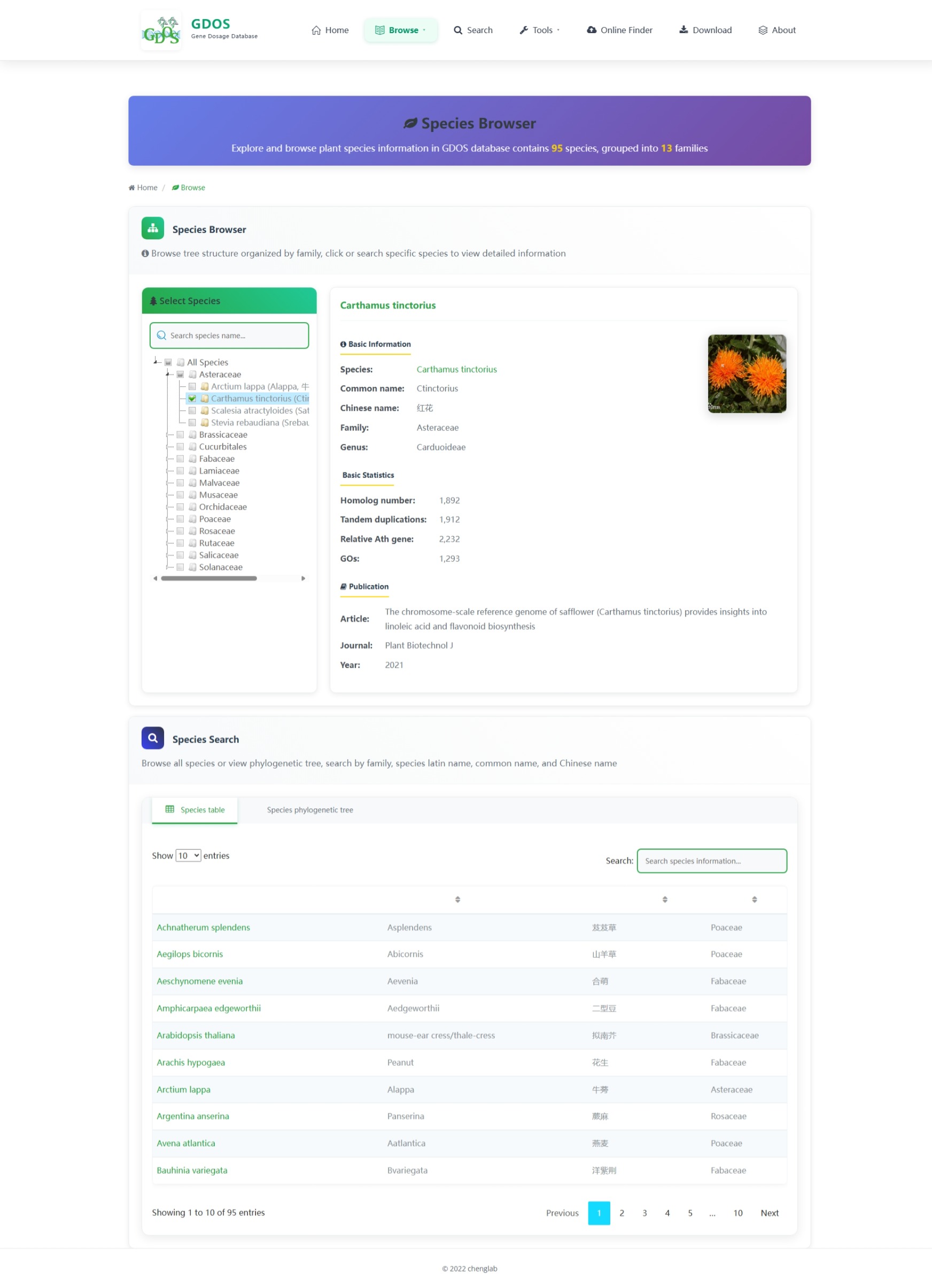
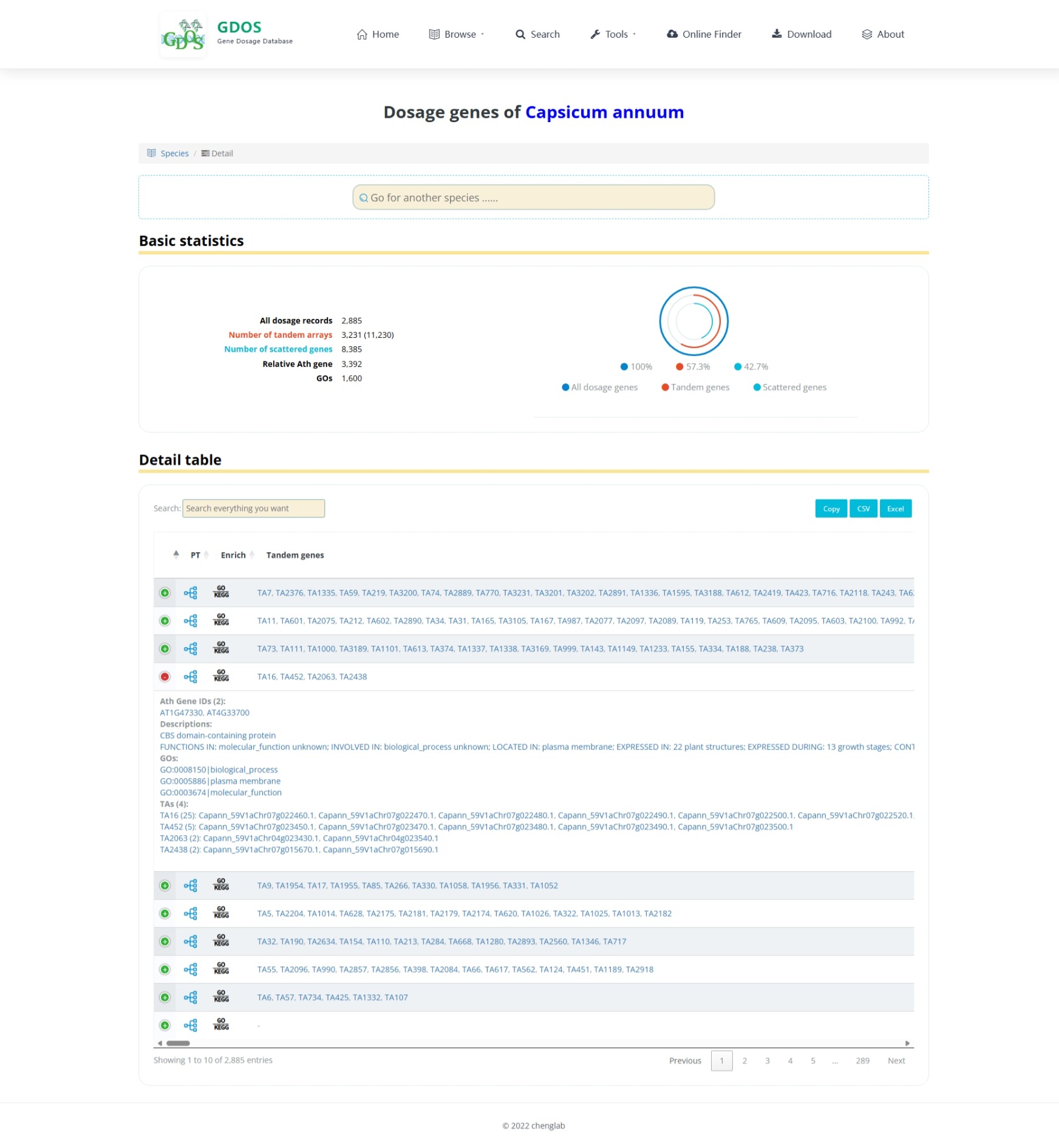
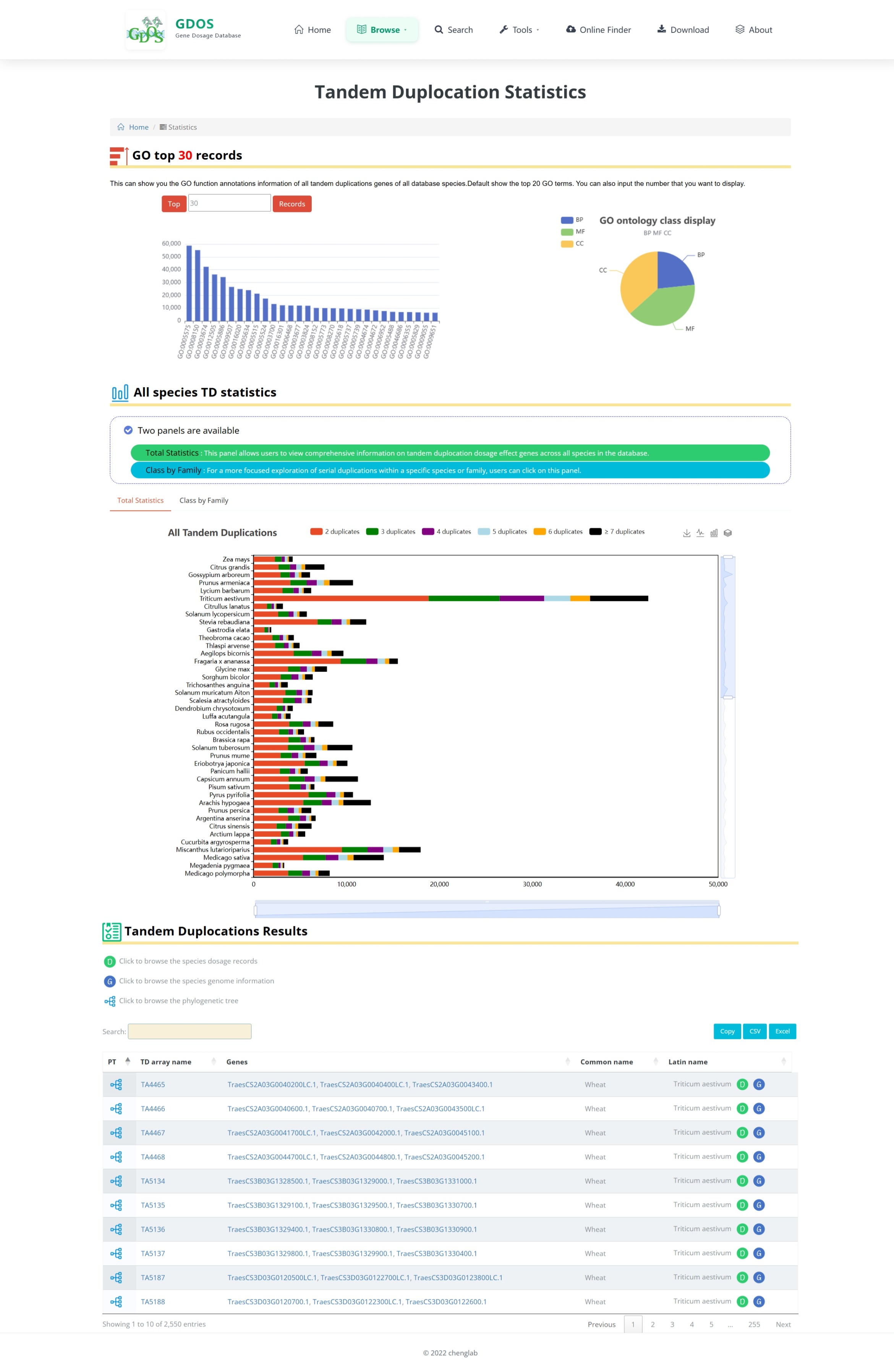
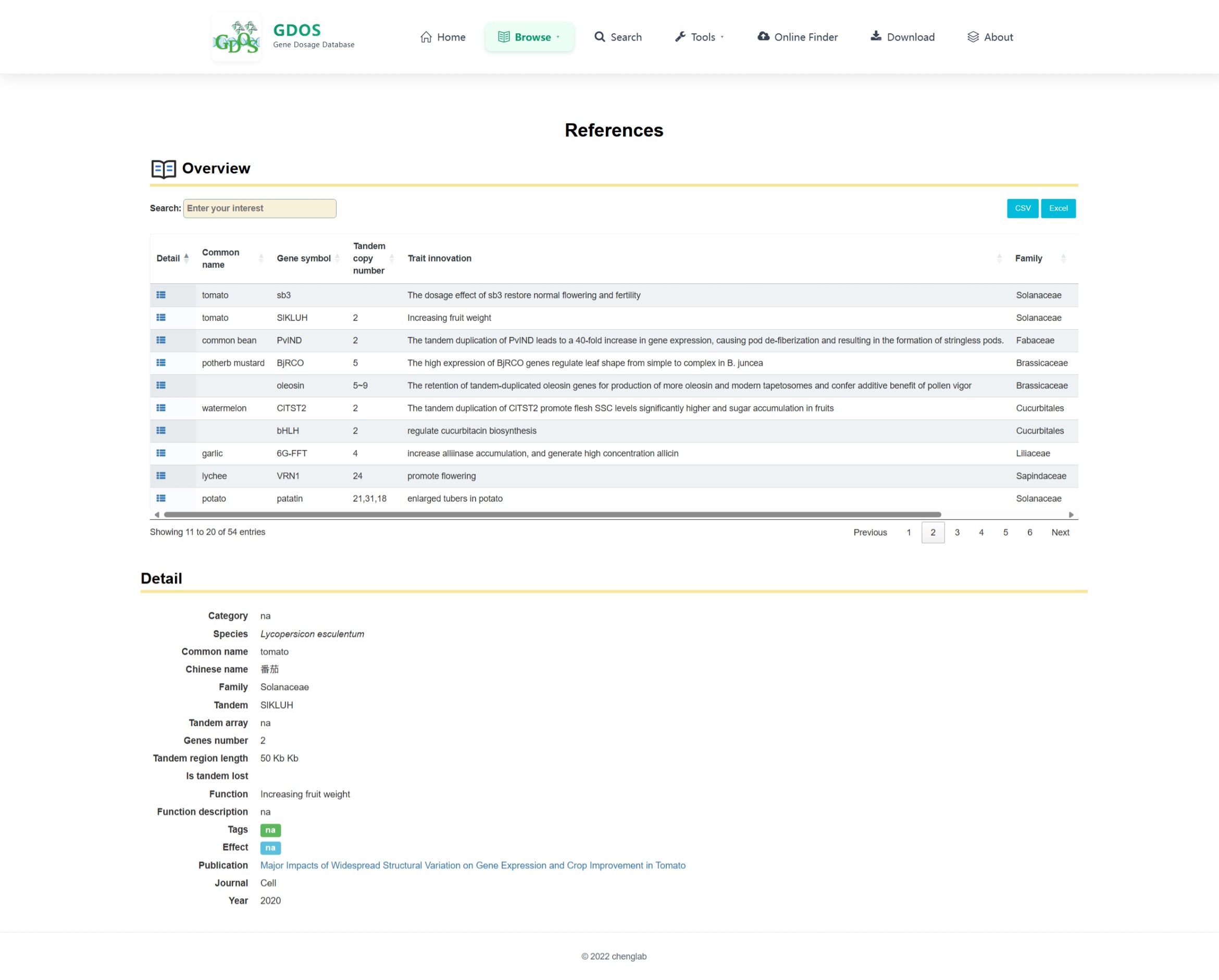
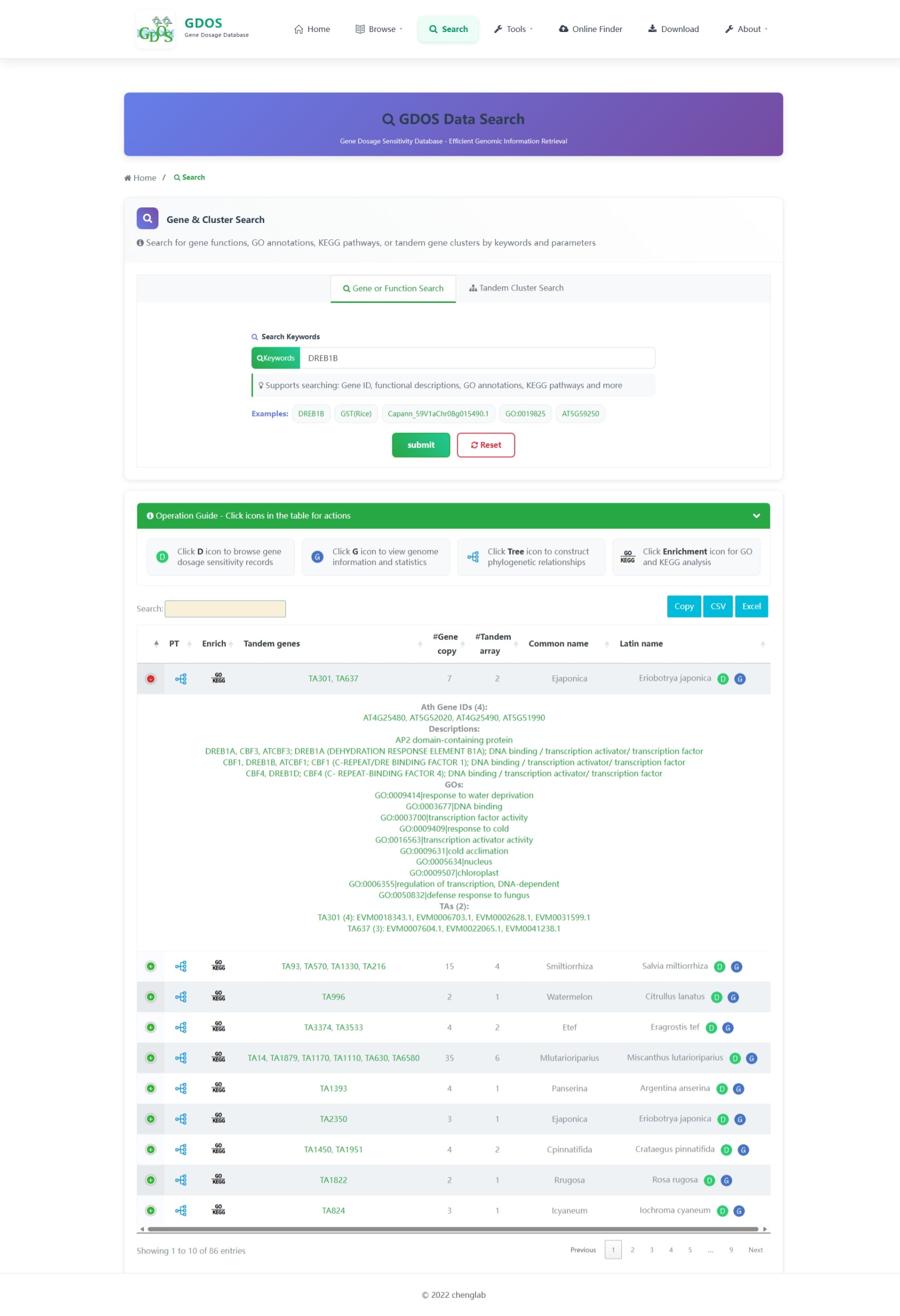
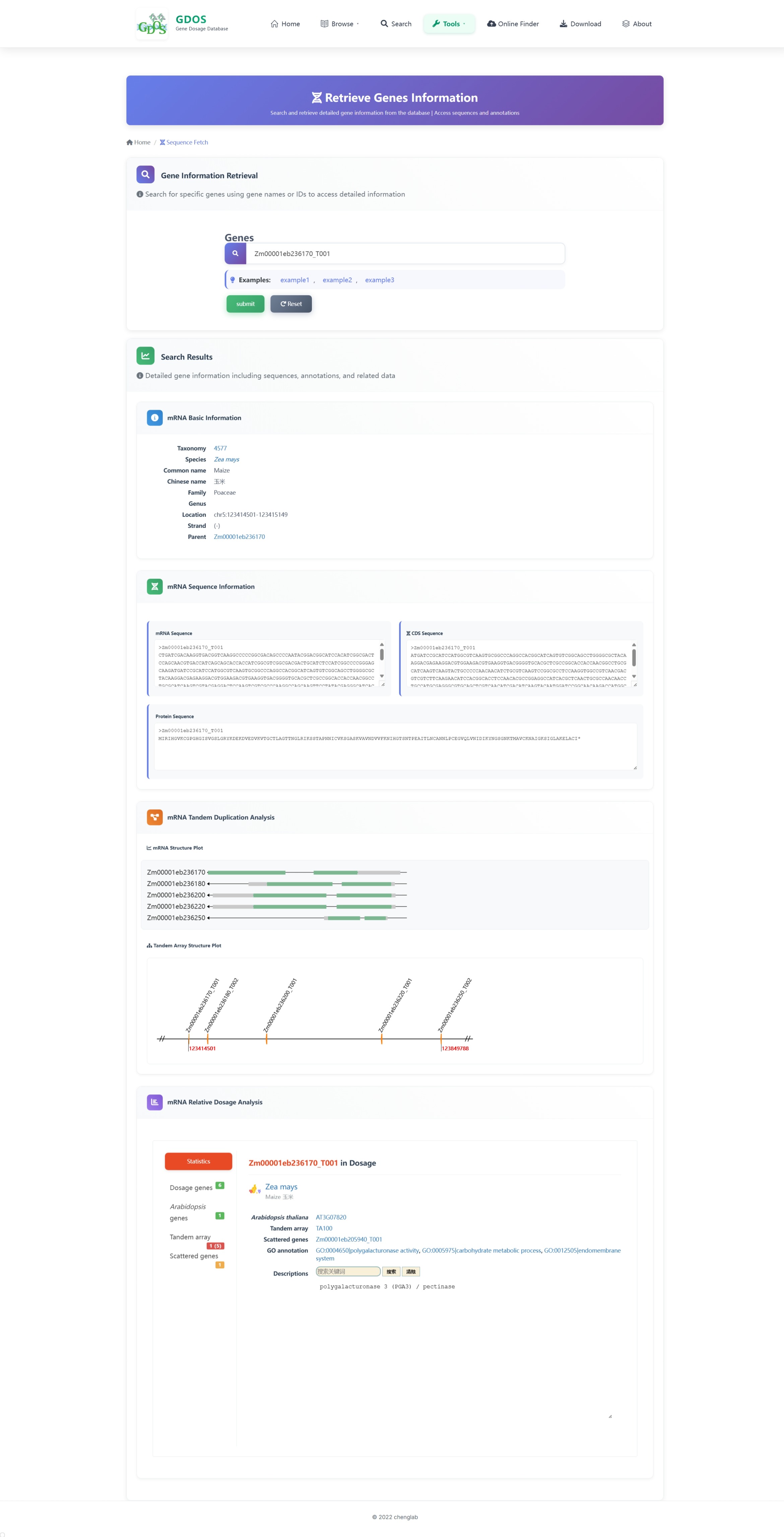
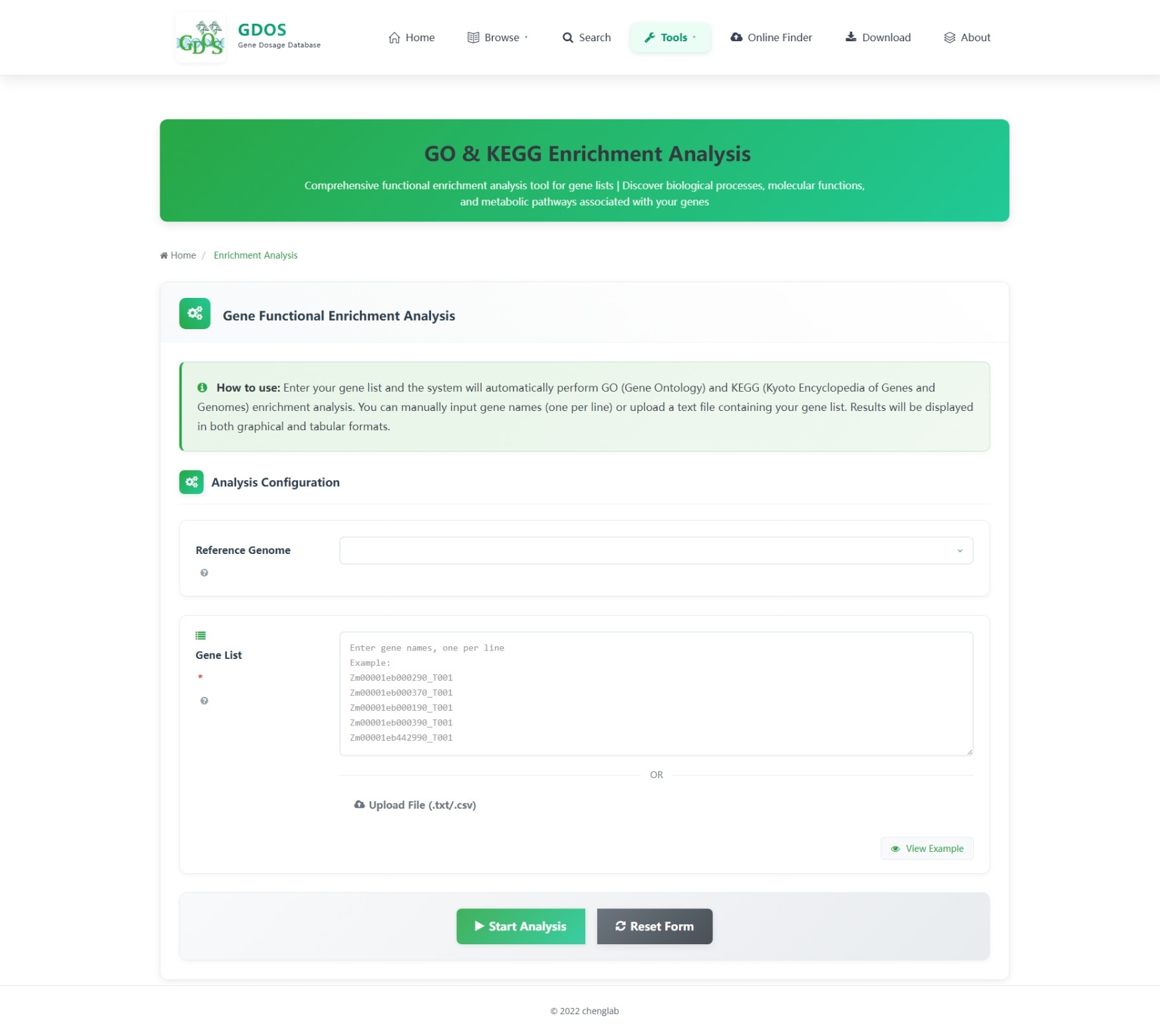
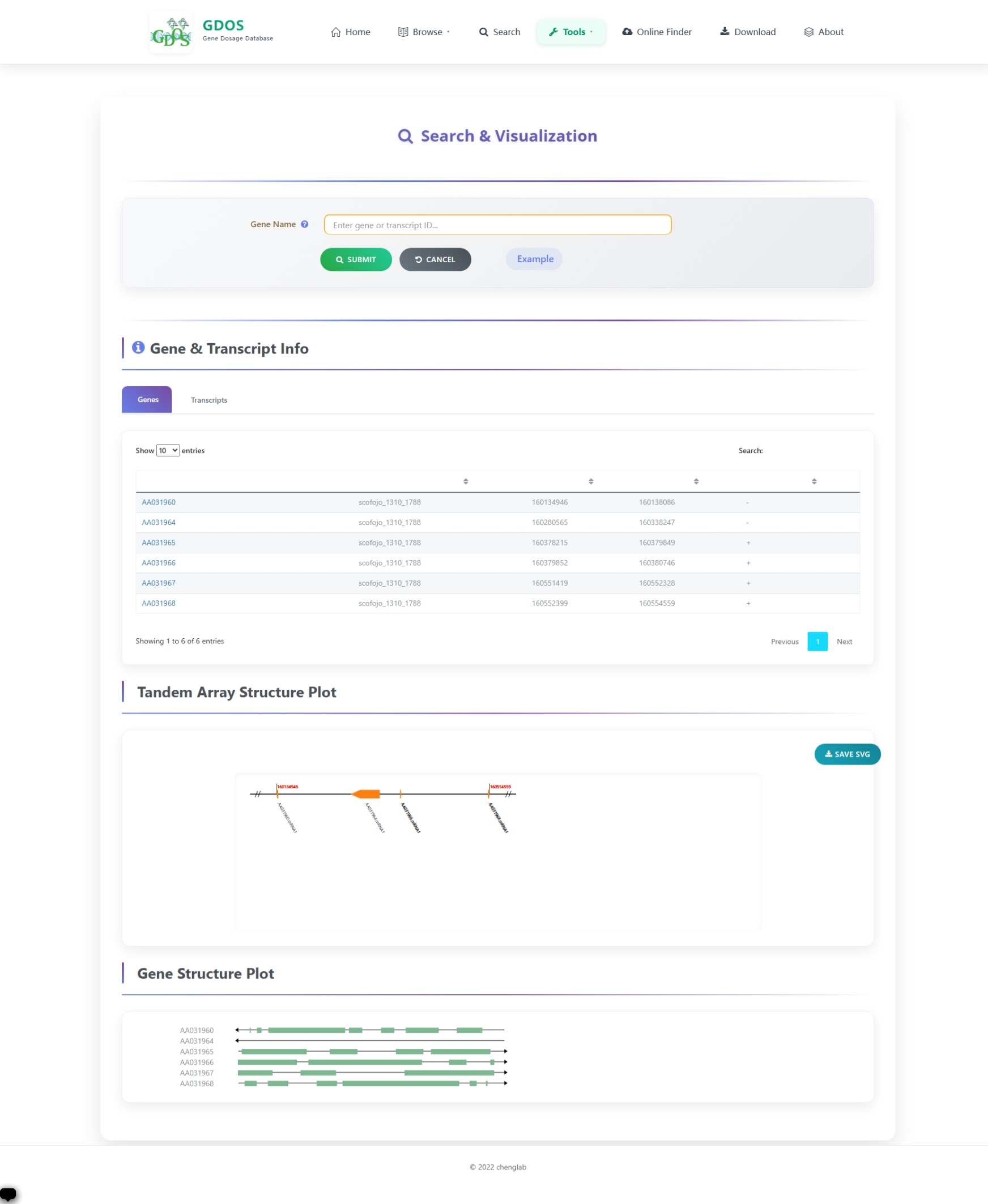
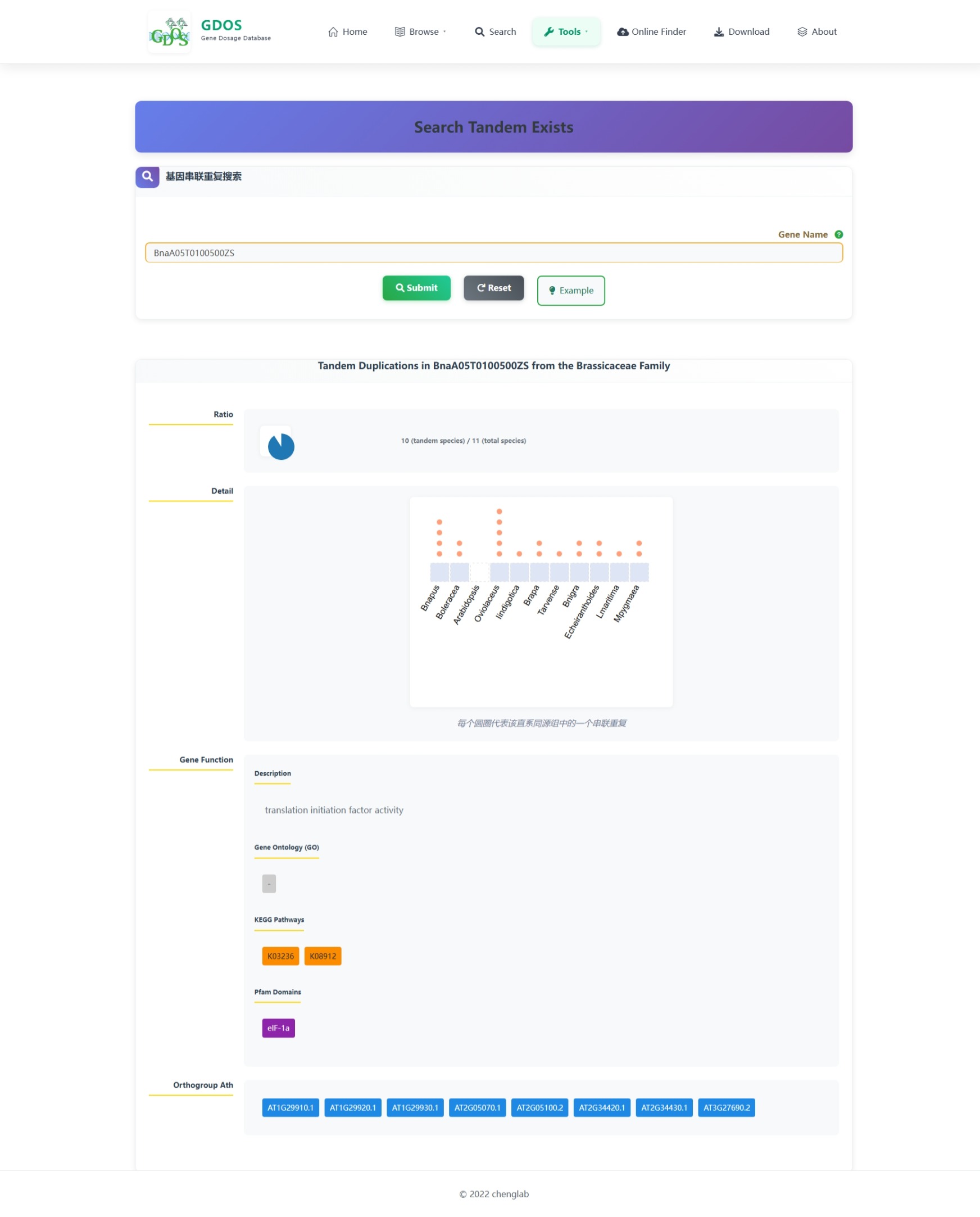
The homepage search engine is a real-time intelligent retrieval system that supports various input types, including gene IDs, Arabidopsis thaliana IDs, GO terms, and functional descriptions. The minimum query string length is 2 characters. For example, to search for fruit ripening-related content, users can input "Frui" (4 characters), and the interface will instantly return the most relevant matches. After selecting the desired entry, pressing Enter twice will execute the search and automatically redirect to the results page.